Concepts¶
Stimulus Synchronization in Neurophysiology¶
Researchers focused on sensation and perception in neuroscience often need high temporal precision in when images or videos are rendered, beyond the abilities of most consumer-grade equipment. To know with millisecond precision when an image was displayed on a computer requires taking into account details like monitor refresh rate, CPU and memory usage, eliminating background tasks in the operating system, etc. to have a reproducible delay between the experimenter hitting start and the stimulus being displayed. Even still, there may be dropped frames.
Alternatively, one can allow for the above sources of variation, but instead record the outcome and piece together timing after the fact. The Element takes this approach by dedicating a corner of the screen to displaying a unique sequence. When captured by a photosensitive diode and decoded, this sequence provides the exact timing for each frame of the stimulus regardless of dropped frames or real-time operating system inaccuracies.
Key Partnerships¶
- Andreas Tolias Lab (Baylor College of Medicine)
Element Roadmap¶
Element Visual Stimulus is a self-contained application that generates and presents visual stimuli using Psychtoolbox, as well as records conditions and trials in a DataJoint database. Further development of this Element is community driven. Upon user requests and based on guidance from the Scientific Steering Group we will continue adding features to this Element.
- Set parameters related to the display of gratings, dots, or 'trippy' (i.e. black and white psychedelic gradient) stimuli.
- During presentation, the corner of the screen is reserved for displaying a photodiode, which provides the exact timing for each frame of the stimulus.
- While Element Visual stimulus is MATLAB-native, the resulting data can be retrieved in Python as part of a larger workflow. For information on running MATLAB scripts with Python, see MathWorks documentation.
- Integration with Element Event
Element Architecture¶
Each of the DataJoint Elements creates a set of tables for common neuroscience data modalities to organize, preprocess, and analyze data. Each node in the following diagram is a table within the Element or a table connected to the Element.
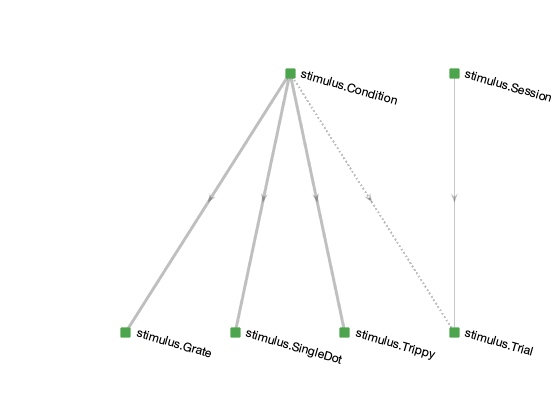
Condition¶
The central table is stimulus.Condition, which enumerates all possible stimulus
conditions to be presented. It is populated before the stimulus is presented for the
first time. The specialization tables below it contain parameters that are specific to
each type of stimulus. For example, stimulus.Monet2 contains parameters that are
specific to a single stimulus condition of the type Monet2. For each tuple in
stimulus.Condition, exactly one of the specialization tables contains the
corresponding entry. The name of the specialization table is indicated in each row of
stimulus.Condition in field stimulus_type.
Example data:
| CONDITION_HASH | stimulus_type | stimulus_version |
|---|---|---|
| +9mOEvwZHyV2MiwRBsMy | stimulus.Varma | 1 |
| +eFINMa+jF58wHzuk9qQ | stimulus.Monet | 1 |
| +0cObnxIHpoB5RKZJVYj | stimulus.Matisse | 1 |
| +9nMtSVLIPAj/VEmey+6 | stimulus.Matisse | 2 |
| +cI6EqAdQgh2tyJ1eMzy | stimulus.Matisse | 2 |
Trial¶
The table stimulus.Trial contains the information about the presentation of a
condition during a specific scan (from experiment.Scan). Any number of conditions of
any type can be presented during a scan and each condition may be displayed multiple
times.
Example data:
| ANIMAL_ID | SESSION | SCAN_IDX | TRIAL_IDX | condition_hash | last_flip | trial_ts | flip_times |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | Qjz5gJN2igKvsonApHO1 | 21322 | 2022-04-21 16:23:40 | =BLOB= |
| 0 | 0 | 0 | 1 | KMk2le1nd79vP4uhW+lG | 21324 | 2022-04-21 16:23:42 | =BLOB= |
| 0 | 0 | 0 | 2 | d3TMSkOO74Y2QzRngY9r | 21325 | 2022-04-21 16:23:43 | =BLOB= |